Research
Sub: Cancer Genetics Research
Tumor microenvironment research HKUMed, 01/2021-present
Summary: Developing computational methods to study cancer genomes (WGS/WES/RNAseq/scRNAseq/Spatial transcriptomics, with particular interest in the interplay between an evolving cancer and a dynamic immune microenvironment.
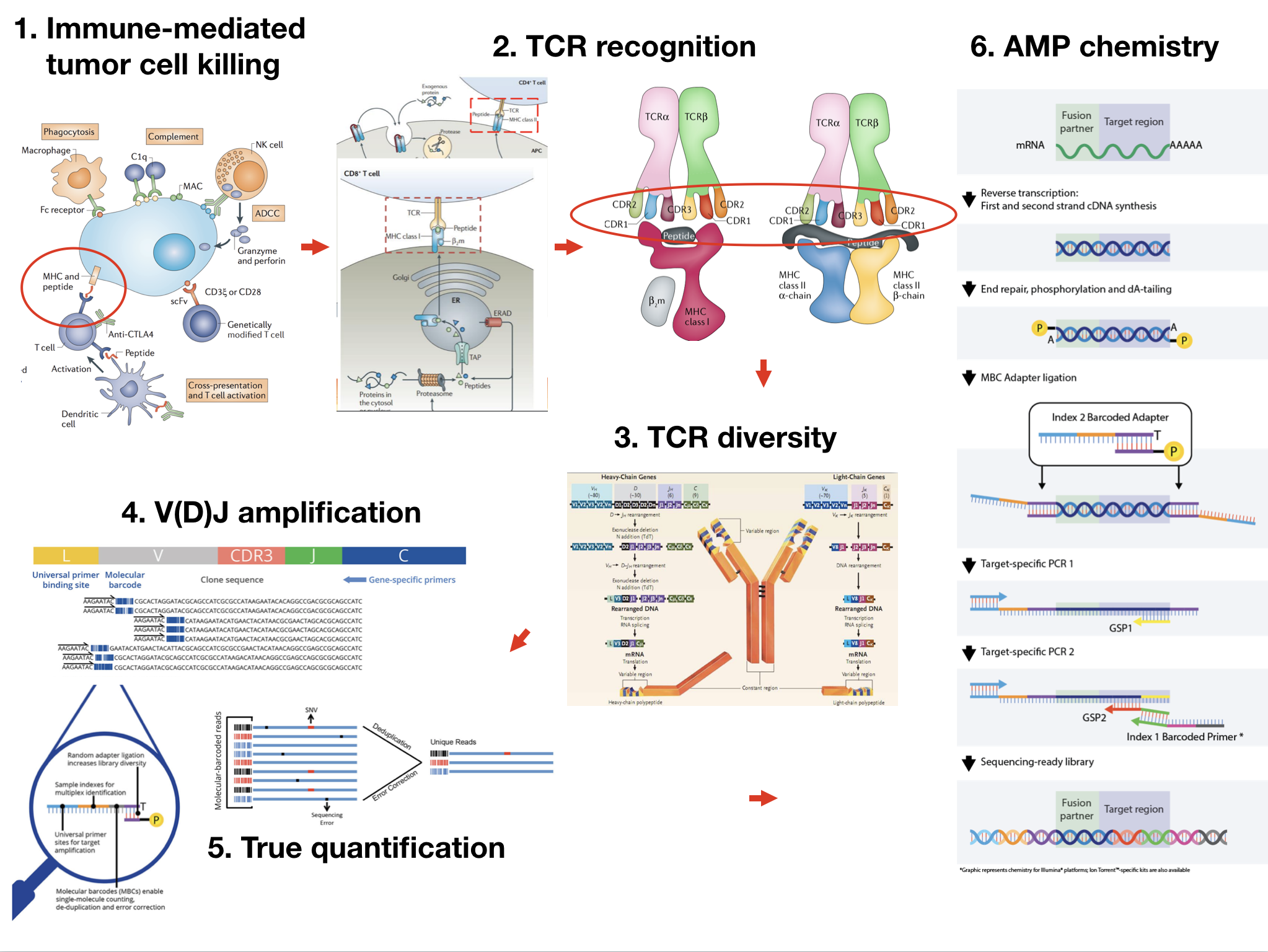
A utra-sensitive and fast platform for the large-scale clinical screening of oncogenic fusions Karolinska Institutet, 04/2018-04/2020
Summary: Wrote code and paper. A companion data analysis platform named SplitFusion, to detect gene fusion based on split alignments, such as reads crossing fusion breakpoints, with the ability to accurately infer in-frame or out-of-frame of fusion partners of a given fusion candidate. SplitFusion also outputs example breakpoint-supporting sequences in FASTA format, allowing for further investigations. SplitFusion can economically and effectively distinguish true oncogenic fusions from environment noise, amplification or sequencing errors, especially for low quality input of clinical samples.
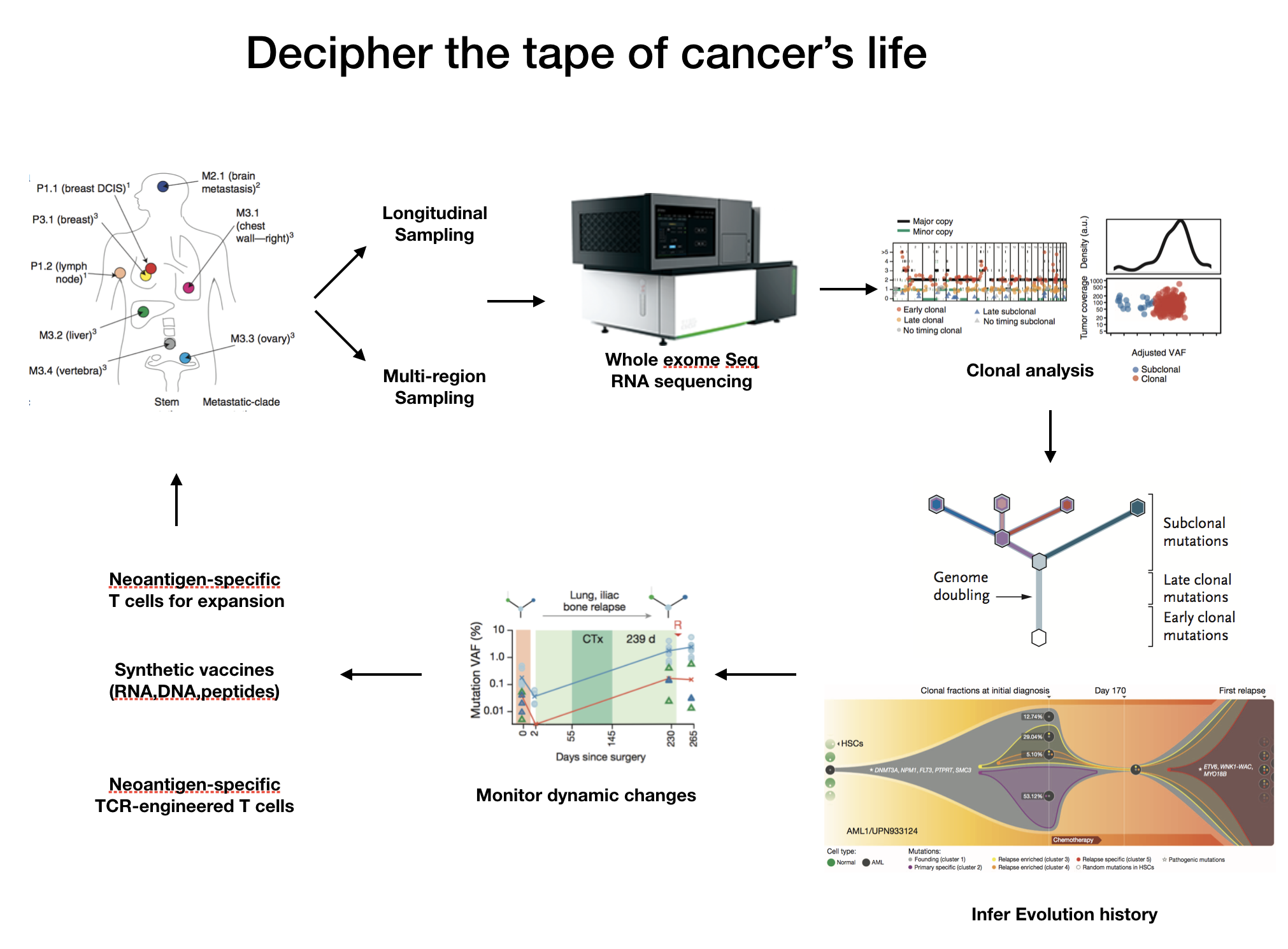
Deciphering genetic clonal diversity and evolution history of clear cell renal cell carcinomas BGI-Shenzhen, 05/2014-04/2018
Summary: Administered and involved in all aspects of the project, wrote the paper independently. Responsible for the clonal evolution structure and immunogenicity analysis of 489 clear cell renal carcinomas patients from Japanese/Chinese/TCGA cohorts, including mutational signature analysis, intra-tumor heterogeneity and cancer evolution history analysis, tumor specific neo-antigen analysis, survival analysis.
Renal cancer genome study by whole genome sequencing of 61 tumor-normal pairs BGI-Shenzhen, 05/2014-04/2018
Summary: Responsible for the personalized analysis of the project including the detection of somatic structure variants, false positive results filter, SV cluster chain identification, result interpretation, and pan-cancer integrated analysis of TCGA database.
Ovarian cancer genome study by Reduced representation bisulfite sequencing and whole transcriptome sequencing of 21 tumor-normal pairs BGI-Shenzhen, 05/2014-04/2018
Summary: Administered and involved in all aspects of the project. Responsible for the detection of differentially methylated regions (DMRs) and DMRs annotation, the correlation analysis between DMRs and gene expression/splice site/transcript factor site, to study the effect of epigenetic modifications in ovarian cancer tumorigenesis.
Nasopharyngeal Carcinoma molecular subtyping study based on somatic mutation and copy number variations BGI-shenzhen 05/2014-04/2018
Summary: Applied network-based stratification (NBS) method in stratification of Nasopharyngeal Carcinoma subtypes by clustering together patients with mutations in similar network regions, extracted molecular feature from NPC subtypes, identified some subtypes that are predictive of clinical outcomes such as patient survival.
Sub: Population Genetics Research
Whole-Genome Resequencing of 330 indigenous inhabitants living along the Tibetan Yi Corridor BGI-shenzhen 08/2013-04/2014 Summary: Responsible for whole process of data mining including raw data filter, quality control, reads alignment, variants detection, variants annotation, data statistics.
Sub: Molecular Biology
Development of assay for detecting the potential microcystin-producing Microcystis in the aquatic ecosystem NBU 07/2012-08/2013
Summary: Designed a new loop-mediated isothermal amplification (LAMP) assay based on microcystin biosynthesis genes. Administered and involved in all aspects of the project including project designing, assay development, result interpretation, and wrote the paper independently.
Screening Adenylylation Domain of NRPS Genes in Marine Bacterial NBU 03/2011-06/2012
Summary: Designed the PCR primer, Performed Molecular cloning experiment including Bacterial culture, Constructed vector, connection, conversion, coated plates, select monoclonal colony PCR.
Work experience
Research Assistant at Ming Wai Lau Centre for Reparative Medicine, Karolinska Institutet, Hong Kong (04/2018 – 04/2020)
Bioinformatics Engineer at Department of Cancer Genetics Research, BGI-Shenzhen, Shenzhen, China (08/2013 – 04/2018)