Future motivations
As for the plan of future period, B. Zhang will focus on three research directions below:
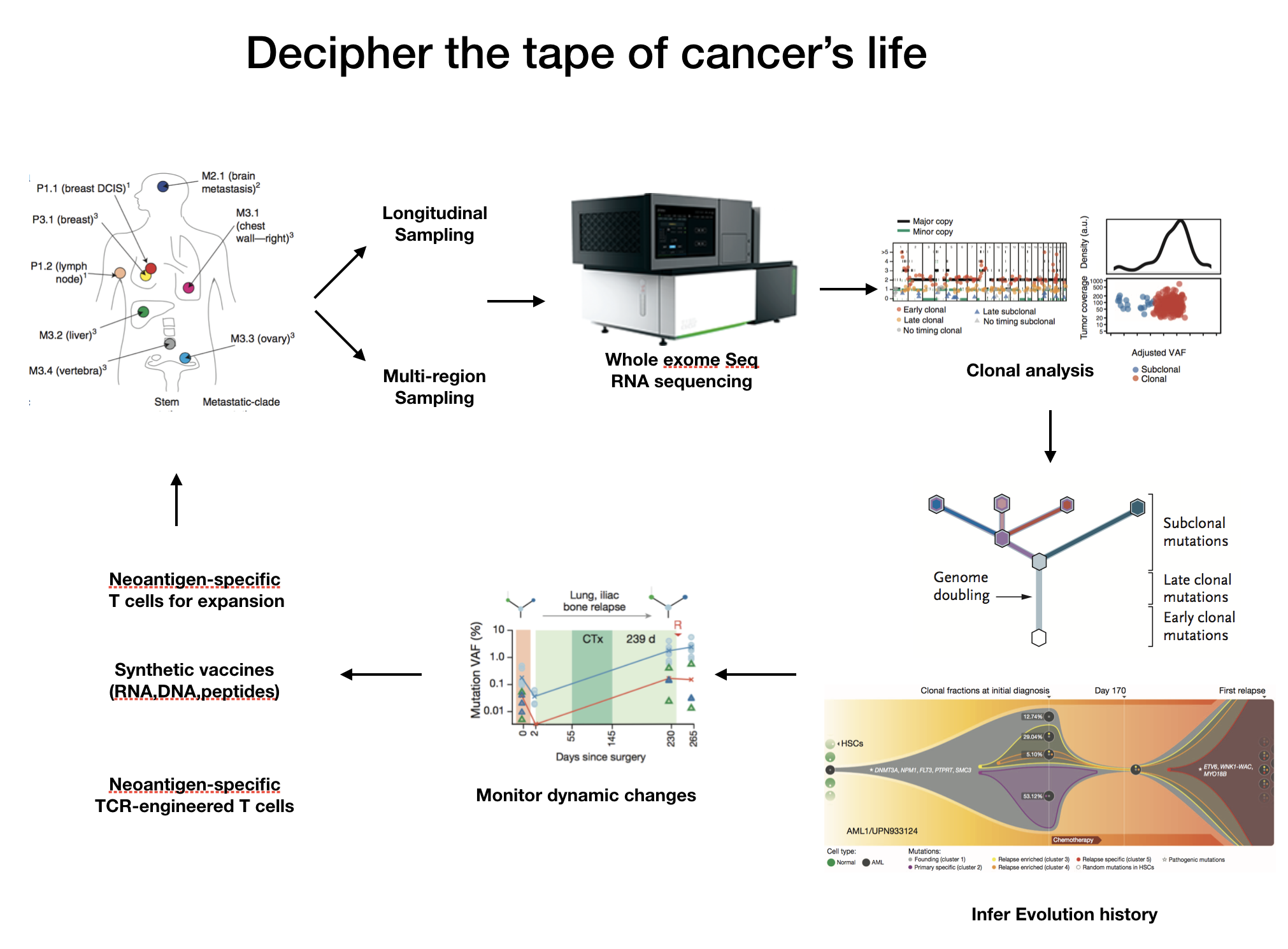
Cancer Evolution
Deciphering dynamic clonal evolution of tumor and monitoring their genomic alterations underlying drug treatment through ctDNA sequencing and analysis. With the innovation in sequencing technology and exponentially decreasing cost of NGS sequencing, hundreds of thousands of somatic mutations involved in tumorigenesis have been identified. Due to the existence of intra or inter tumor heterogeneity, simply aggrandizing sequencing project scale to uncover more or rare novel variants can’t help us to beat cancer more efficiently. B. Zhang realize that the most important thing we need to focus on is to understand the role of mutations played in tumor evolution and their relation with patients survival outcome, which could give us more precise direction to personalized therapy and to evaluate the treatment efficacy.
Cancer Immunology
Exploiting vaccines binding to tumor-specific neo-antigen to elicit immunoreactivity in cancer patients. Thousands of mutation offers abundant fuels of vaccines, especially which in clonal state to some extent has higher priority in drug development and can be developed to expand the repertoire of neoantigen-specific T cells and to induce patients immunoreactivity. The use of personal vaccines can address tumor heterogeneity as well as minimize the chance of tumor escape by loss of antigen.
Deep learning
Mining of tumor genomics database and applying deep learning algorithms to perform molecular subtyping. There are several well-established cancer databases such as TCGA and ICGC, which incubated multi-omics data of about 30 tumor type. These databases have detailed clinical information including clinical outcome, treatment response and drug response. B. Zhang wants to apply many optimized deep learning framework such as Tensorflow/caffe in extracting clinical related molecular feature from multilayered omics data, and to build some sensitive and specific subtyping model to stratify cancer patients into distinct group. For example, to develop radiomics signature corresponded to molecular subgroup, which may serve as a complementary noninvasive tool for tumor genetic test.